Common Optogenetic Proteins
The optogenetic toolbox used in non-neuronal optogenetics consists of a variety of photoactivatable proteins and their interaction partners. Generally speaking, the underlying mechanism of most of these proteins is light-dependent monomerization or oligomerization. Until today, the range of optogenetic tools is constantly being expanded, either by genetically modifying and optimizing existing optogenetic proteins or by discovering entirely novel proteins or protein domains. They originate from all three domains of life and can thus be found in Archaea (such as halorhodopsin), Bacteria (such as bacteriophytochrome), and Eukaryota (such as channelrhodopsin).
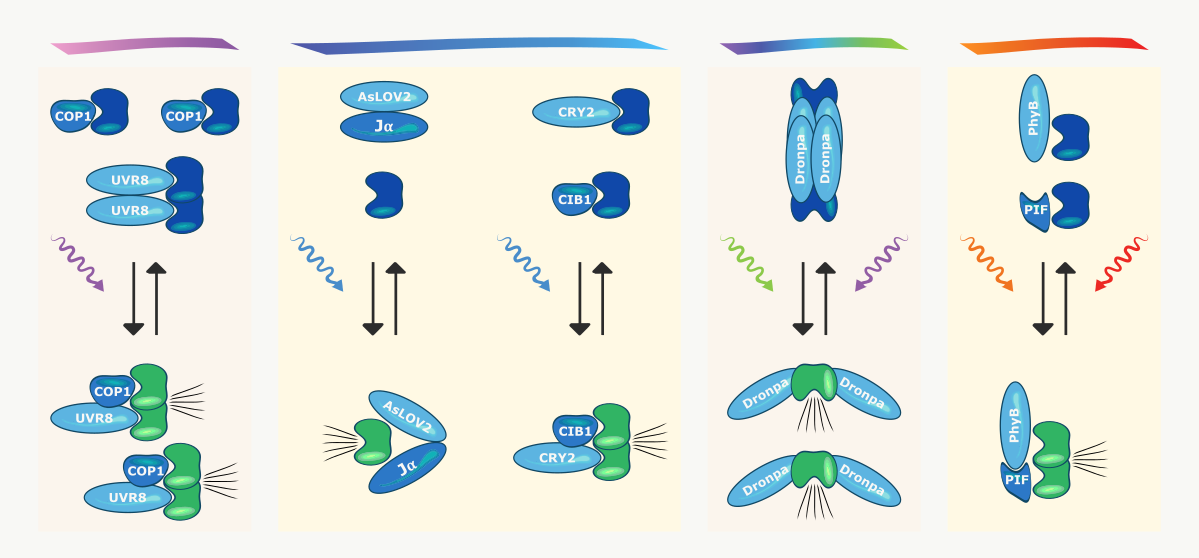
The commonly used plant UV photoreceptor UVR8 forms a stable homodimer in the dark. Ultraviolet irradiation induces rapid disruption and dissociation of the dimer. Monomeric UVR8 is then able to bind to its interaction partner COP1, which activates the function of the fused protein of interest and its downstream signaling cascade. Relaxation of the system is achieved in the dark and can take several hours.
Another optogenetic tool found in plants is the flavin mononucleotide binding protein AsLOV2 with its C-terminal Jα alpha helix. Blue light irradiation causes the Jα alpha helix to swing out from the LOV core domain, revealing a previously hidden binding site for a downstream signaling protein. Photoreversion in the dark occurs on a timescale of minutes.
Blue light illumination of the plant cryptochrome CRY2 leads to photoactivation and subsequent binding to its interaction partner CIB1. Similar to the UVR8-COP1 system, this triggers a downstream signaling cascade from the fused protein of interest. Within minutes, the CRY2-CIB1 heterodimers dissociate in the dark, terminating the signal.
Among the optogenetic proteins, Dronpa occupies a special role. Not only can it reversibly switch between a tetrameric and a monomeric state by blue/green and violet light, respectively. Beyond that, it is also a switchable fluorescent protein, exhibiting strong fluorescence only in its oligomeric state. Dronpa has been used to cage fusion protein function in its oligomeric state. Monomerization after illumination exposes the active sites of these proteins of interest, which activates their initial cellular function.
Lastly, phytochromes are red/far-red light-responsive photoreceptors found in microbes and plants. The widely used PhyB/PIF optogenetic system from the model plant Arabidopsis thaliana makes use of the light-dependent interaction of the phytochrome PhyB with the phytochrome interacting factor PIF. Thus, fusion to output domains is used to reliably switch between active and inactive signaling states in the range of milliseconds to seconds.